Glucagon receptor crystal structure reveals class B GPCR signal transduction mechanisms
— Complex structure of a class B GPCR bound to an analogue of its endogenous ligand ignites new excitement in GPCR research
A team of scientists from Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences has determined the high-resolution atomic structure of the human glucagon receptor (GCGR), which plays a key role in glucose homeostasis, in complex with a peptide analogue of the endogenous ligand glucagon. This structure reveals, for the first time, the molecular details of a class B GPCR binding to its peptide ligand at high resolution and unexpectedly discloses the structural complexity that governs receptor activation, thereby greatly expanding our understanding of class B GPCR signal transduction.
In a paper published in Nature on January 4th, 2018 (London time) entitled “Structure of the glucagon receptor in complex with a glucagon analogue” (DOI: 10.1038/nature25153), scientists at SIMM, in collaboration with several groups based in China (Fudan University, iHuman Institute at ShanghaiTech University, Zhengzhou University and East China Normal University), Denmark (Novo Nordisk), Canada (University of Toronto) and United States (GPCR Consortium), provided a detailed molecular map of the full-length GCGR in complex with a glucagon analogue and partial agonist NNC1702.
Class B GPCRs exert essential action in hormonal homeostasis and are important therapeutic targets for a variety of diseases, including metabolic disorders such as type 2 diabetes, osteoporosis, migraine, depression and anxiety. These receptors consist of an extracellular domain (ECD) and a transmembrane domain (TMD), both of which are required to interact with their cognate peptide ligands and to regulate downstream signal transduction. Due to difficulties in high-quality protein preparation, structure determination of full-length class B GPCRs remains challenging, thus limiting a comprehensive understanding of molecular mechanisms of receptor action.
Activation of GCGR by glucagon triggers the release of glucose from the liver during fasting, and thus it is a potential drug target for type 2 diabetes. Last year, the scientists at SIMM determined the crystal structure of the full-length GCGR bound to a negative allosteric modulator NNC0640 and an inhibitory antibody mAb1, which, for the first time, provides a clear picture of a full-length class B GPCR at high resolution. To better understand how class B GPCRs recognize cell signals, the same group of scientists recently made further progresses by solving the crystal structure of GCGR in complex with the glucagon analogue NNC1702. According to team leader and SIMM professor Dr. Beili Wu, “The newly solved GCGR structure not only helps us understand the signal recognition mechanisms of class B GPCRs, but also provides the most accurate template to date for drug design targeting GCGR, which offers new opportunities in drug discovery for treating type 2 diabetes.”
This study gives some valuable insights into the activation mechanism of GCGR. The most exciting finding is that the linker region connecting the ECD and TMD of the receptor, termed the “stalk”, and the first extracellular loop undergo significant conformational changes in their secondary structures in the peptide-bound GCGR structure compared to the previously determined non-peptide-bound structure. This leads to a marked change in the relative orientation between the ECD and TMD of the receptor to accommodate peptide binding and initiate receptor activation. Furthermore, the stalk may modulate receptor activity by facilitating conformational movements of the receptor TMD. “It is amazing to observe how the stalk region plays such an important role in regulating receptor function, although it only contains 12 amino acids,” said Dr. Qiang Zhao, the co-corresponding author of the paper. “This has never been observed in previously solved GPCR structural studies. It significantly deepens our knowledge about class B GPCR signaling mechanisms.”
Based on the structure of GCGR-NNC1702 complex, the researchers performed a series of functional studies using techniques such as competitive ligand binding, cell signaling, molecular dynamics simulations and double electron-electron resonance spectroscopy. The results are in support of the GCGR structure and confirm the conformational alterations of the receptor in different functional states. This study was supported by various research teams from SIMM, Fudan University, iHuman Institute at ShanghaiTech University, University of Toronto and Novo Nordisk. “This is achieved by a team effort with experts from different fields, indicating that Shanghai has become a GPCR research center of excellence that will bring more opportunities in international collaborations in the future,” said Dr. Hualiang Jiang, Director of SIMM.
In addition to Drs. Wu, Zhao and Jiang, other notable investigators included Dr. Ming-Wei Wang (holding a concurrent professorship at Fudan University), Dr. Dehua Yang and graduate student Haonan Zhang (the first author) from SIMM, Dr. Raymond Stevens from the iHuman Institute, ShanghaiTech University, Dr. Steffen Reedtz-Runge and Dr. Jesper Lau from Novo Nordisk, Dr. Oliver Ernst and Dr. Ned Van Eps from University of Toronto, and Dr. Michael Hanson of GPCR Consortium. The study was funded by Chinese Academy of Sciences, the National Science Foundation of China, Shanghai Science and Technology Development Fund, Shanghai Municipal Education Commission and the Canada Excellence Research Chairs program.
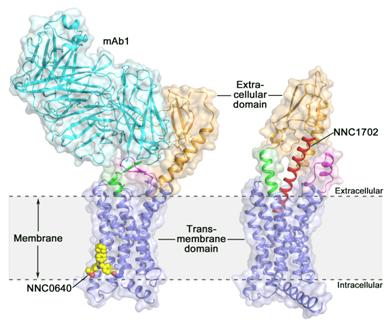
The new Nature study reports the crystal structure of the full-length human glucagon receptor (GCGR), which plays a key role in glucose homeostasis and serves as an important drug target for type 2 diabetes, in complex with an analogue of its endogenous ligand glucagon. The image shows the overall architecture of the GCGR-NNC1702 complex (right) and the previously determined crystal structure of GCGR bound to a negative allosteric modulator NNC0640 and an antibody mAb1 (left). The structures are colored orange (extracellular domain), blue (transmembrane domain), green (stalk), magenta (the first extracellular loop), red (NNC1702), yellow (NNC0640) and cyan (mAb1). (Image courtesy of Dr. Beili Wu.)
